class: center, middle  <!-- Do not forget to adapt the presentation title in the header! --> <!-- Adjust the presentation to the session. Focus on the challenges, this is not a coding tutorial. Note, to include figures, store the image in the `/docs/assets/images` folder and use the jekyll base.url reference as done in this template or see https://jekyllrb.com/docs/liquid/tags/#links. using the scale attribute , you can adjust the image size. --> <!-- Adjust the day, month --> # 23 MAY 2019 ## INBO coding club <!-- Adjust the room number and name --> Herman Teirlinck<br> 01.71 - Frans Breziers --- class: center, middle <!-- Create a new badge using Inkscape based on the assets/images/coding_club_badges.svg file -->  Special thanks to our database admins Jo Loos and Gert Van Spaendonk! --- class: center, middle 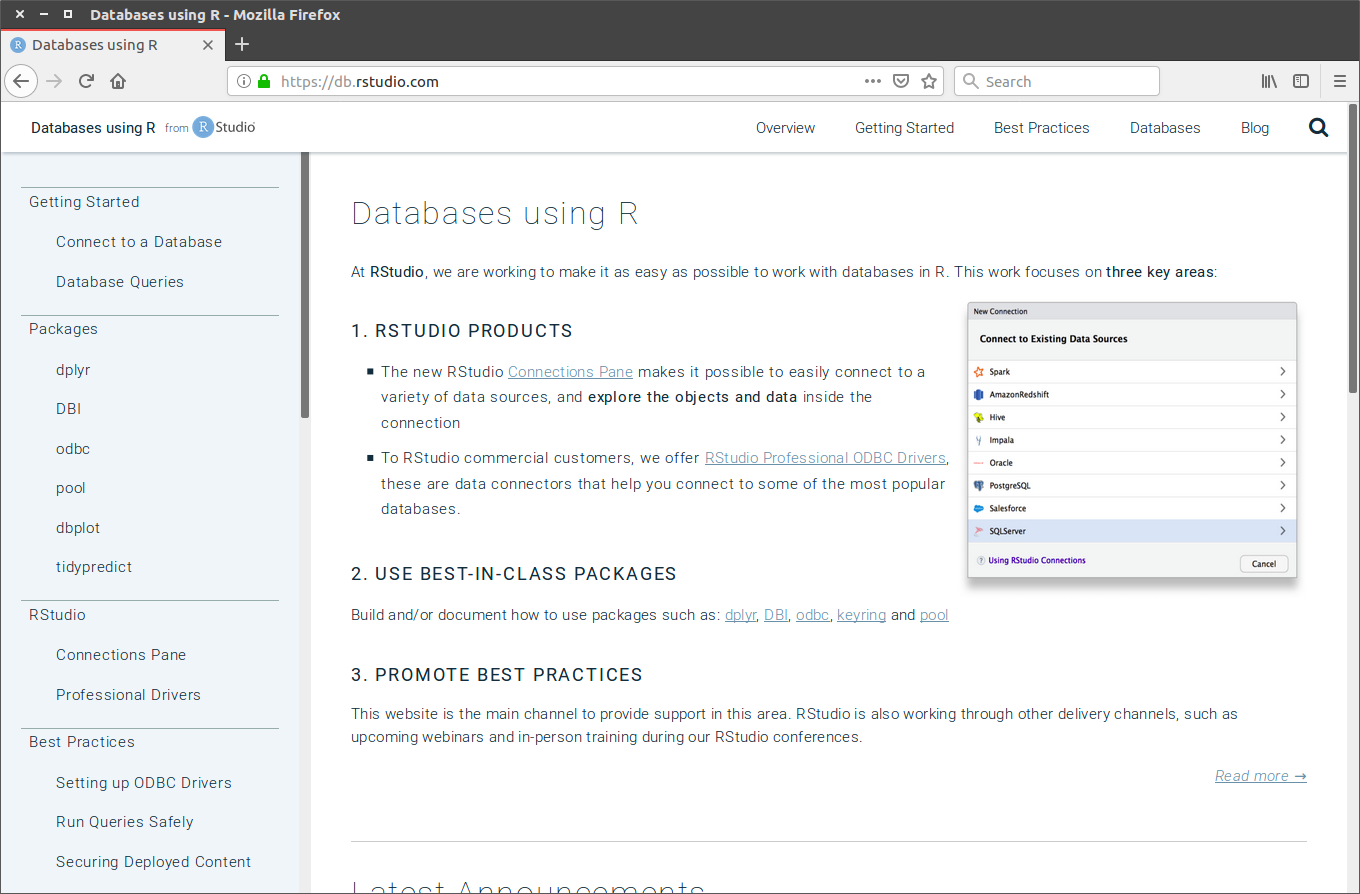<br /> [Rstudio database website](https://db.rstudio.com/) --- class: center, middle 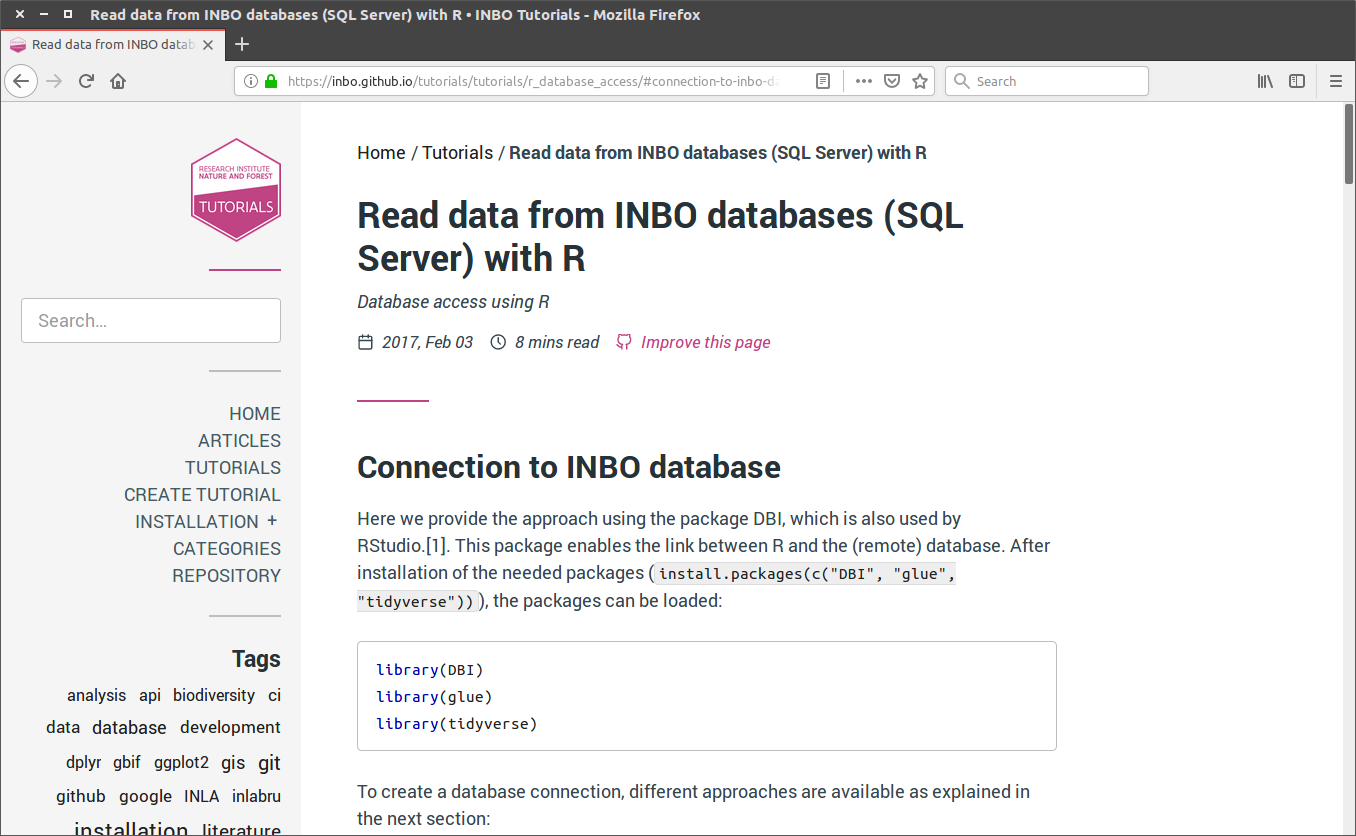<br /> [INBO database tutorial](https://inbo.github.io/tutorials/tutorials/r_database_access/) --- class: center, middle ### How to get started? Check the [Each session setup](https://inbo.github.io/coding-club/gettingstarted.html#each-session-setup) to get started. ### First time coding club? Check the [First time setup](https://inbo.github.io/coding-club/gettingstarted.html#first-time-setup) section to setup. --- class: center, middle 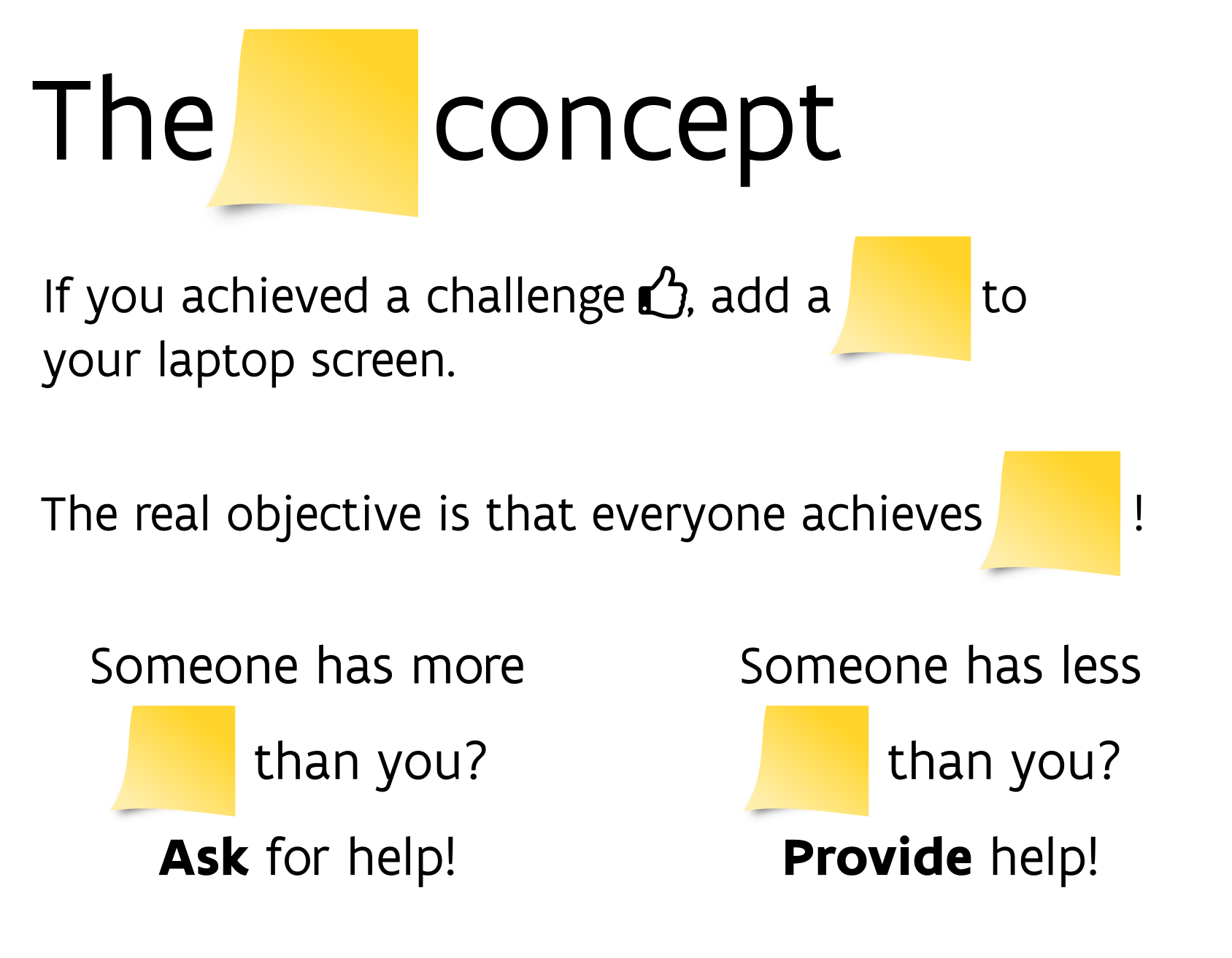 --- class: center, middle ### Share your code during the coding session! <!-- Create a new hackmd file and replace this link (twice!) --> Go to https://hackmd.io/pcYIBNEJScGDEeIvc9LOEg <iframe src="https://hackmd.io/pcYIBNEJScGDEeIvc9LOEg" height="400px" width="800px"></iframe> --- background-image: url(/assets/images/background_challenge_1.png) class: left, middle # Challenge 1 Using the [INBO database tutorial](https://inbo.github.io/tutorials/tutorials/r_database_access/): - Click [`20190523/20190523_challenges.R`](https://github.com/inbo/coding-club/blob/master/src/20190523/20190523_challenges.R) and download* script to `src` subfolder. - Install and load the [inborutils](https://inbo.github.io/inborutils/index.html) package - Use the [`connect_inbo_dbase`](https://inbo.github.io/inborutils/reference/connect_inbo_dbase.html) function from the `inborutils` package to make a connection to the "W0003_00_Lims" or the "D0021_00_userFlora" database - Can you get an overview of the tables in the database using the Rstudio interface? <small>* __Note__: check the getting started instructions on [how to download a single file](https://inbo.github.io/coding-club/gettingstarted.html#each-session-setup)</small> <br><br><br> <small>Thanks to Floris for reviewing and improving the database tutorial and database connection function yesterday!</small> --- class: left, middle ## Intermezzo: start to SQL... - `SELECT`: which __columns__ are you interested in? - `FROM`: which __table__ are you interested in? - `TOP N`: __limit__ to the top N rows, with N equal to a number - `DISTINCT`: __unique__ values within a given column - `WHERE`: __filter__ the data with specific conditions __REMEMBER:__ `TOP 10` is your friend, keep traffic low! Use `SELECT/WHERE` to limit if possible <br><br> <small>Want to dive a bit deeper? <br> Check the [Data carpenty: data Management with SQL for Ecologists](https://datacarpentry.org/sql-ecology-lesson/) lesson, especially on [queries](https://datacarpentry.org/sql-ecology-lesson/01-sql-basic-queries/index.html) and [aggregations](https://datacarpentry.org/sql-ecology-lesson/02-sql-aggregation/index.html). </small> --- background-image: url(/assets/images/background_challenge_2.png) class: left, middle # Challenge 2 The LIMS database `W0003_00_Lims` contains the labo measurements with an important table `FactResult`. We want to have more insight into the column names of the `FactResult` table and will setup [a query](https://inbo.github.io/tutorials/tutorials/r_database_access/#execute-a-query-to-the-database) for this: - Make a connection to the database `W0003_00_Lims`. - Get first 10 rows of the table `vwDocumentatie` with a query. - Create a query to get the unique values of the `name` column. - Query from table `vwDocumentatie` the documentation by filtering those rows where the `name` is equal to 'Description'. - Store the output of the query to a new object called `fact_columns_documentation`. --- background-image: url(/assets/images/background_challenge_3.png) class: left, middle # Challenge 3 Writing queries can be hard and we have specialists in house who are great at it... Let's make sure we can reuse our own and their hard work! We have a custom query available to get the scientificname of 'Gebogen kransblad': ``` custom_query <- " SELECT NaamWetenschappelijk FROM dbo.tblTaxon WHERE NaamNederlands = 'Gebogen kransblad' " ``` Create a function called `userflora_query_scientificname`: - The function has two input arguments: a connection and a Dutch species name (_not just 'Gebogen kransblad', any UserFlora name_) - The function returns a dataframe with the corresponding scientificname of UserFlora - Test your function by using it, <br>e.g. `userflora_query_scientificname(con, 'Mijtermos')` __Note:__ [This](https://inbo.github.io/tutorials/tutorials/r_database_access/#create-and-use-query-templates) part of the tutorial can be useful... --- class: center, middle <!-- Always try to include some type of reviewing (can be short and entertaining). Remove this slide if no reviewing is included -->  --- class: center, middle 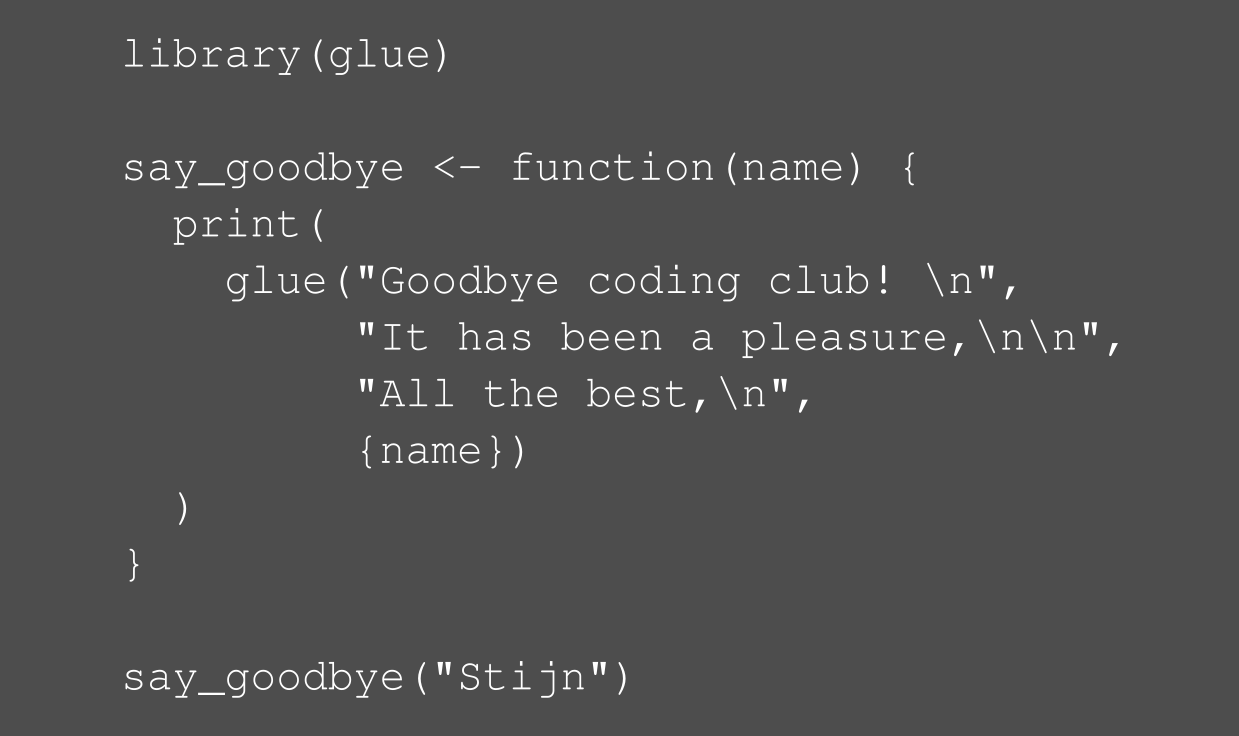 --- class: center, middle  <!-- Adjust the room and date --> Room: Herman Teirlinck - 01.71 - Frans Breziers<br> Date: __28/06/2019__, van 10:00 tot 12:00<br> (registration announced via DG_useR@inbo.be)