class: center, middle  <!-- Do not forget to adapt the presentation title in the header! --> <!-- Adjust the presentation to the session. Focus on the challenges, this is not a coding tutorial. Note, to include figures, store the image in the `/docs/assets/images/yyyymmdd/` folder and use the jekyll base.url reference as done in this template or see https://jekyllrb.com/docs/liquid/tags/#links. using the scale attribute , you can adjust the image size. --> <!-- Adjust the day, month --> # 25 APRIL 2023 ## INBO coding club <!-- Adjust the room number and name --> Herman Teirlinck Building 01.05 - Isala Van Diest --- class: center, middle <!-- Create a new badge using Inkscape or other programs based on the assets/images/coding_club_badges.svg file -->  --- class: left, top ### How to get started? Check the [Each session setup](https://inbo.github.io/coding-club/gettingstarted.html#each-session-setup) to get started. ### First time coding club? Check the [First time setup](https://inbo.github.io/coding-club/gettingstarted.html#first-time-setup) section to setup. --- class: left, top  --- class: center, top # Share your code during the coding session <!-- Create a new hackmd file and replace this link (twice!) --> Go to https://hackmd.io/peUHuQaERrCi94LObScfdA?edit and start by adding your name in section "Participants". <iframe src="https://hackmd.io/peUHuQaERrCi94LObScfdA?edit" height="400px" width="800px"></iframe> --- class: left, top # Download data and code You can download the material of today: - automatically via `inborutils::setup_codingclub_session()`* - manually** from GitHub folders [coding-club/data/20230425](https://github.com/inbo/coding-club/tree/master/data/20230425) and [coding-club/src/20230425](https://github.com/inbo/coding-club/tree/master/src/20230425) <br> <small> __\* Note__: you can use the date in "YYYYMMDD" format to download the coding club material of a specific day, e.g. run `setup_codingclub_session("20220428")` to download the coding club material of April, 28 2022. If date is omitted, i.e. `setup_codingclub_session()`, the date of today is used. For all options, check the [tutorial online](https://inbo.github.io/tutorials/tutorials/r_setup_codingclub_session/).</small> <br> <small> __\*\* Note__: check the getting started instructions on [how to download a single file](https://inbo.github.io/coding-club/gettingstarted.html#each-session-setup)</small> --- class: left, top # Data and scripts description Today we will work with national/regional checklist data about Invasive Alien Species (IAS) and species distribution of butterflies among several biotopes. - `20230425_ias_first_observed_BE.txt`: dataset about year of first observation of IAS in Belgium from 1950 - `20230425_ias_first_observed_regions.txt`: dataset about year of first observation of IAS in Belgian regions from 1950 - `20230425_ias_pathways_count.txt`: dataset about pathway of introduction of IAS in Belgium from 1950 - `20230425_area_biotopes.txt`: dataset about biotopes and distribution of two butterflies - `20230425_challenges.R`: R script with code to start with --- class: left, top # Load libraries ```r library(tidyverse) library(patchwork) library(ggforce) library(gganimate) library(plotly) ``` --- class: left, top # RECAP: ggplot recipe: data - mapping - geometry 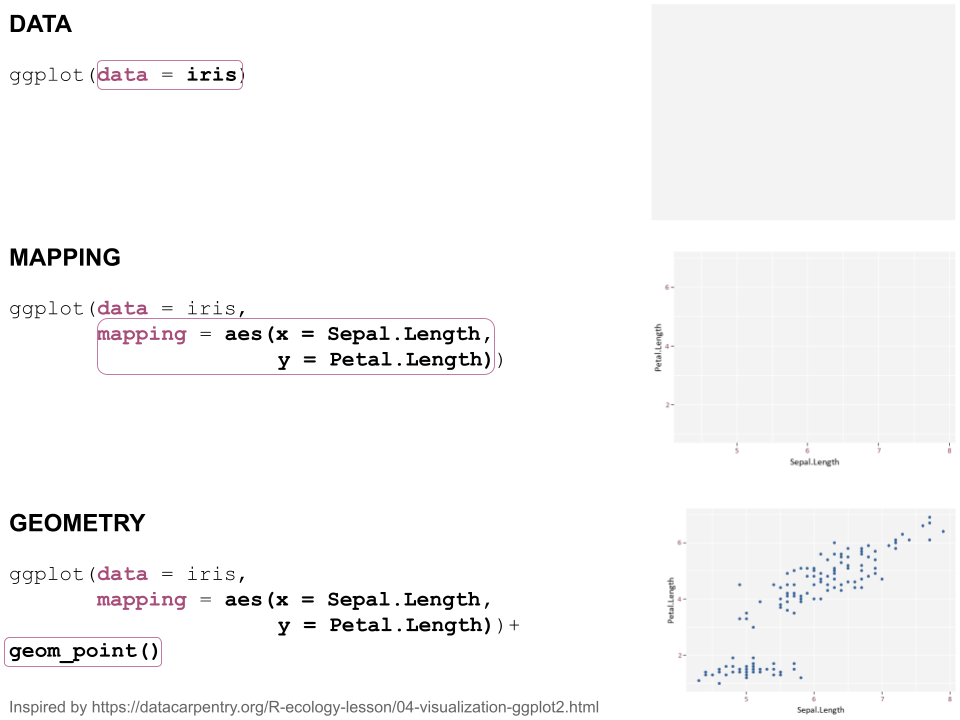 --- class: left, top ## Recap: mapping Mapping can be defined in either `ggplot()` or the geometry `geom_*()`. ```r # mapping in ggplot() ggplot(ToothGrowth, mapping = aes(x = dose, y = len)) + geom_point() # mapping in geometry geom_point() ggplot(ToothGrowth) + geom_point(mapping = aes(x = dose, y = len)) ``` What to do with multiple plots (geometries)? ```r # set all args of aes() in each geom_*() ggplot(ToothGrowth) + geom_violin(mapping = aes(x = dose, y = len, group = dose)) + geom_point(mapping = aes(x = dose, y = len)) # put common args of aes() in ggplot() ggplot(ToothGrowth, mapping = aes(x = dose, y = len)) + geom_violin(mapping = aes(group = dose)) + geom_point() ``` --- class: left, top ## Recap: mapping - aesthetics The most important mapping argument? **aes()**thetics! Put all arguments which require variables (= columns) in `aes()`, leave all other arguments out. ```r # you want color represents the supplement (supp) ggplot(ToothGrowth, mapping = aes(x = dose, y = len)) + geom_point(aes(color = supp)) # you want all points are blue hollow triangles ggplot(ToothGrowth, mapping = aes(x = dose, y = len)) + geom_point(color = "blue", shape = 2) ``` --- background-image: url(/assets/images/background_challenge_1.png) class: left, top # Challenge 1. Combine separate ggplots Let's play with [patchwork](https://patchwork.data-imaginist.com/), one of the highest rated registered extension of ggplot. 1. Display `ias_first_obs_be` and `ias_first_obs_reg` next to each other 2. Display `ias_first_obs_reg` below `ias_first_obs_be` 3. Similar to 2, but this time instead of showing dodge histograms all together, display different regions as different facets and call it `ias_first_obs_reg_facets`. You should end up with one big histogram on top and 3 histograms (= 3 locations) at the bottom of the plot. Tip: adding a facet to `ias_first_obs_reg` is standard ggplot. 4. Starting from the histogram ias_first_obs_be, display different kingdoms as facets. Then, put this plot on the right, the plots in 3 on the left. 5. Display `ias_first_obs_reg_facets` on the left and a table with three rows showing the total number of introductions at regional level on the right. Hint: check this [vignette](https://patchwork.data-imaginist.com/articles/guides/assembly.html#adding-non-ggplot-content). --- class: left, top # Intermezzo. How to choose a color palette: the viridis palette Today we won't speak about colors. Still, take in mind this criteria when choosing a color palette: 1. printer-friendly (gray scale) 2. perceptually uniform 3. easy to read by those with colorblindness. All of this can be found in the [viridis palette](https://www.r-bloggers.com/2018/07/ggplot2-welcome-viridis/) which avoids the red and goes from blue to yellow, passing through green (_viridis_: green in Latin). More about colors in the Bonus Challenge and Resources slides. --- class: left, top # Intermezzo. The INBO official colors Do you know that INBO has its own (color) theme? It's called [INBOtheme](https://inbo.github.io/INBOtheme/) and it's a package. Install it, load it and all figures will have authomatically an INBO touch. Read the nice [tutorials](https://inbo.github.io/INBOtheme/articles/index.html) for more. ```r library(INBOtheme) ggplot(data = ias_be, mapping = aes(x = first_observed)) + geom_bar() + ggtitle(label = "Temporal distribution of alien species introductions") ``` --- background-image: url(/assets/images/background_challenge_2.png) class: left, top # Challenge 2 We explore a little the extra faceting utilities provided by the [ggforce](https://ggforce.data-imaginist.com/index.html) package. 1. Add a zooming facet to `ias_first_obs_paths` to zoom around the time interval 1980 - 2000. How to zoom on the y-axis values as well? 2. In challenge 1 we used standard ggplot facetting to create `ias_first_obs_reg_facets`. How to use ggforce to set the facet labels (the `locationId` values) on the bottom instead of on the top as usual? --- background-image: url(/assets/images/background_challenge_3.png) class: left, top # Challenge 3A. Animate your plot Using butterflies/biotopes data, create a GIF as the one below by using ggplot2 and [gganimate](https://gganimate.com/) packages. Use `"cubic-in-out"` _easing_ to smooth the transition between states. Save the GIF on disk as well (width = 1000, height = 300). Kind of a bonus: save it as a video instead. 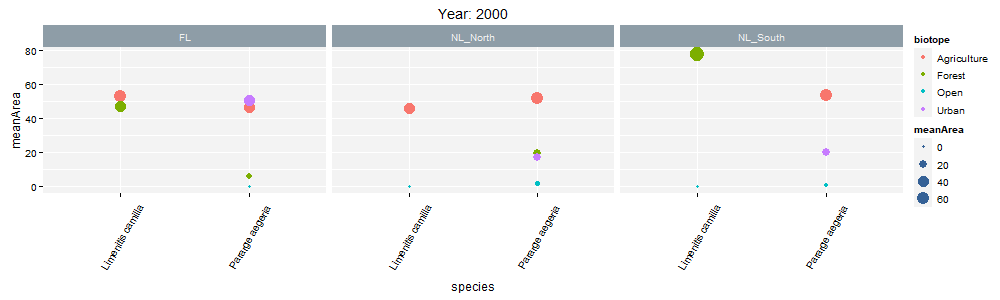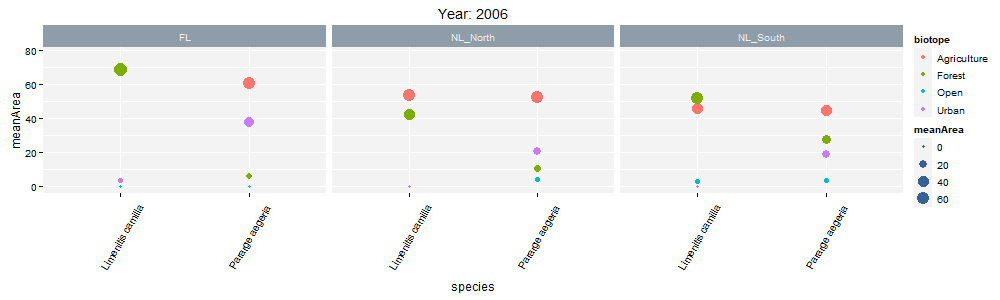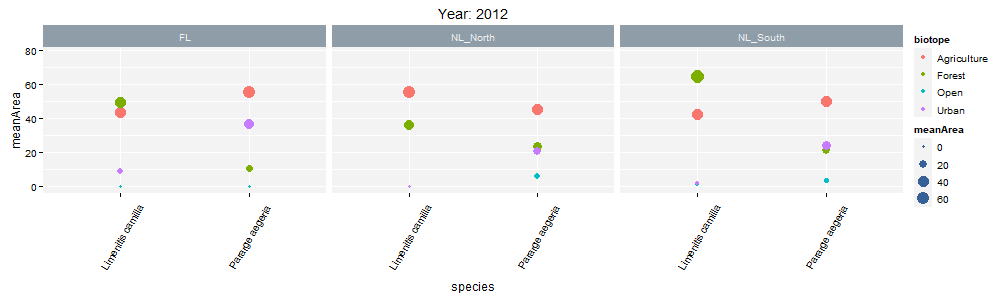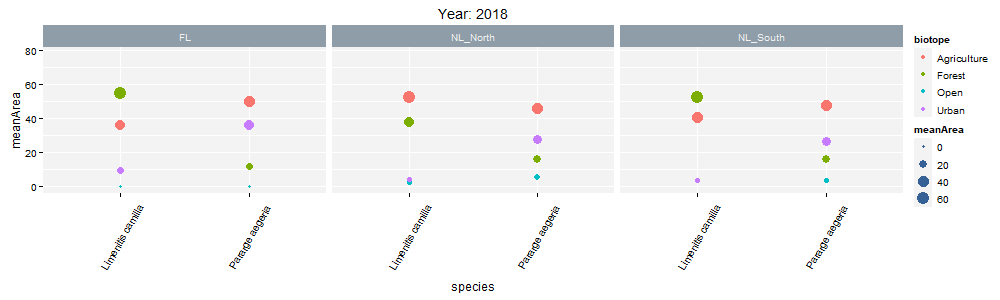 --- background-image: url(/assets/images/background_challenge_3.png) class: left, top # Challenge 3B. Create interactive plots The R package [plotly](https://plotly-r.com/) is an R package for creating interactive web-based graphs via plotly's JavaScript graphing library. In particular, you can make your ggplot plots interactive! 1. Make the plot `ias_first_obs_paths` interactive. Explore the interactivity tools. In particular check the legend filtering capabilities on pathway! Tip: documentation in the ["intro to ggplotly"](https://plotly-r.com/overview.html#intro-ggplotly) section. 2. How to add _dynamic ticks_ functionality to the interactive plot in 1? Dynamic ticks means generating new axis ticks while zooming in/out 3. Improve the text shown while hovering so that it looks something like in screenshot below 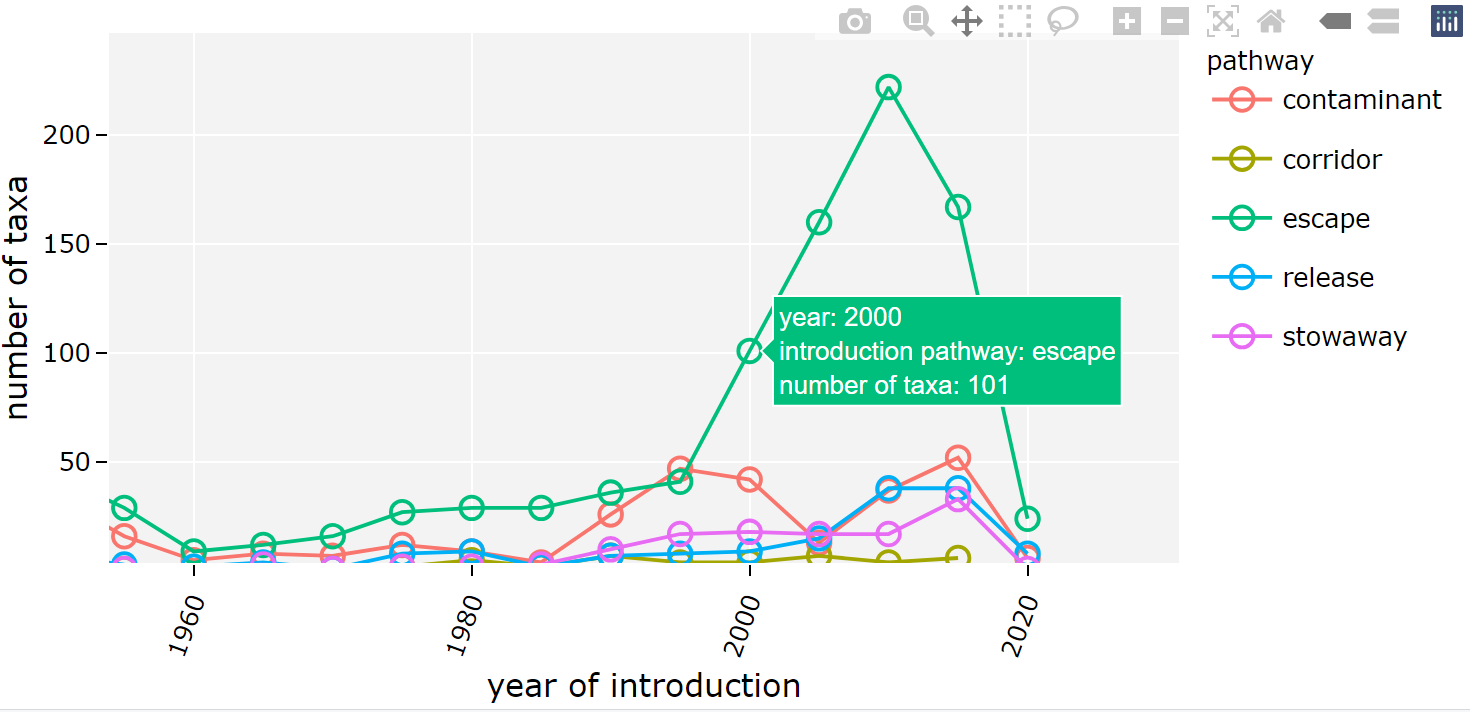 --- class: left, top # Bonus challenge: have fun with color palettes There are so many color palettes available. Get inspired! Examples: - [wesanderson](https://github.com/karthik/wesanderson): it allows you to use (some of) the palettes derived from the amazing Tumblr blog ["Wes Anderson Palettes"](http://wesandersonpalettes.tumblr.com/) - [ggsci](https://nanx.me/ggsci/index.html): it offers a collection of high-quality ggplot compatible color palettes. - [cols4all](https://github.com/mtennekes/cols4all): it is a package to help you selecting color palettes via a GUI (see screenshot in next slide) Try to apply the viridis palette mentioned in the Intermezzo and other palettes from the packages mentioned above to some of the plots we created today. --- class: left, top # Bonus challenge: have fun with color palettes ``` library(cols4all) c4a_gui() # launch the GUI (graphic user interface) ``` 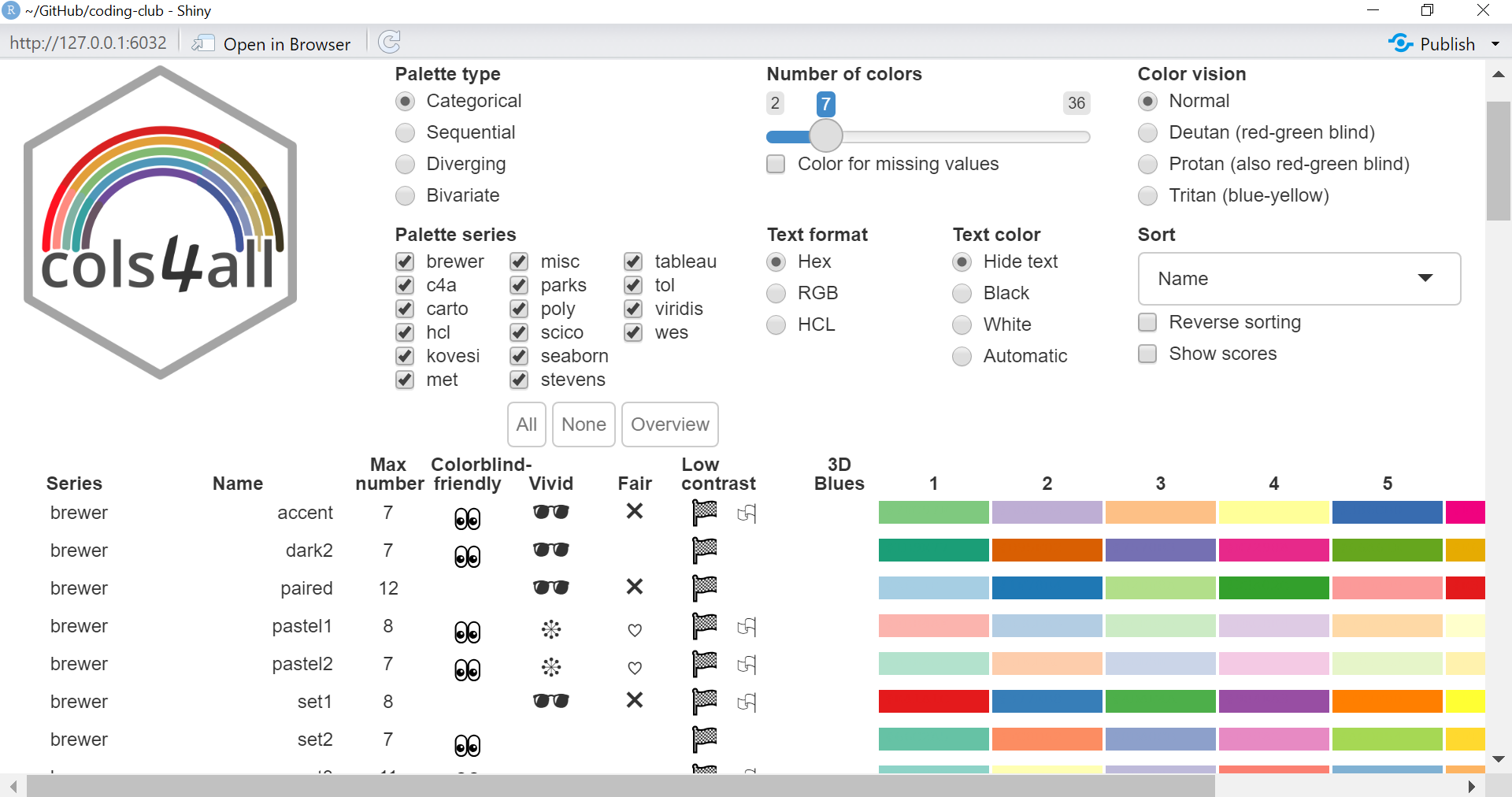 --- class: left, top # The package of the month. Hans' choice 2 Hans Vancalster inspires us again! The [ggpcp](https://yaweige.github.io/ggpcp/index.html) package to create *generalized parallel coordinate plots* in the ggplot2 framework. Interesting also to read [this](https://www.tandfonline.com/doi/full/10.1080/10618600.2023.2195462) article about it. Example:  --- class: left, top # Resources - Commented challenges [solutions](https://github.com/inbo/coding-club/blob/main/src/20230425/20230425_challenges.R) - Edited [video recording](https://vimeo.com/823315912) is available on the vimeo INBO coding club channel - `ggplot2` website: https://ggplot2.tidyverse.org/ - [`ggplot2` extension packages gallery](https://exts.ggplot2.tidyverse.org/gallery/) - Arrange intuitively multiple plots with [patchwork](https://patchwork.data-imaginist.com/) - Explore [ggforce](https://ggforce.data-imaginist.com/): a kind of repository of geoms, stats, etc. that are as well documented and implemented as the official ones found in ggplot2 - Animate your plots with [gganimate](https://gganimate.com/) - Make interactive plots using [plotly](https://plotly-r.com/) - Tutorial about [plotly package](https://www.musgraveanalytics.com/blog/2018/8/24/how-to-make-ggplot2-charts-interactive-with-plotly) for making interactive plots - More about animations? Check [challenge 3B](https://inbo.github.io/coding-club/sessions/20201217_outstanding_plots.html#16) of 17 Dec 2020 - [wesanderson](https://github.com/karthik/wesanderson) packge offers (some of) the palettes shown in the Tumblr blog ["Wes Anderson Palettes"](http://wesandersonpalettes.tumblr.com/) - [ggsci](https://nanx.me/ggsci/index.html) package offers a collection of high-quality ggplot compatible color palettes - [INBOtheme](https://inbo.github.io/INBOtheme/) homepage - Blogpost about colorblindness and how taking care of it in your plots: https://www.datanovia.com/en/blog/how-to-stimulate-colorblindness-vision-in-r-figures - Chapter dedicated to [plotting colors](https://bookdown.org/ndphillips/YaRrr/more-colors.html) from the "YaRrr! The Pirate’s Guide to R" - Rate your visualization using Stephanie Evergreen's [data visualizaion checklist](https://stephanieevergreen.com/data-visualization-checklist/) --- class: left, top # Coding club topics 2023: you vote! Every month you can vote among **two topics**! Poll for May's coding club is still open! Let us know your favorite before May 1st! https://forms.gle/dyBHYSyVWqC1tnk69 You can choose between: - Spatial vector data with sf: points, lines, polygons, multipolygons, ... - The computer says NO: debug yourR code --- class: center, middle  <!-- Adjust the room and date --> Room: 01.20 - Willy Van Der Meeren<br> Date: __25/05/2023__, van 10:00 tot 12:30<br> Subject: "spatial data with sf" or "computer says no: debugging in R"<br> (registration announced via DG_useR@inbo.be)